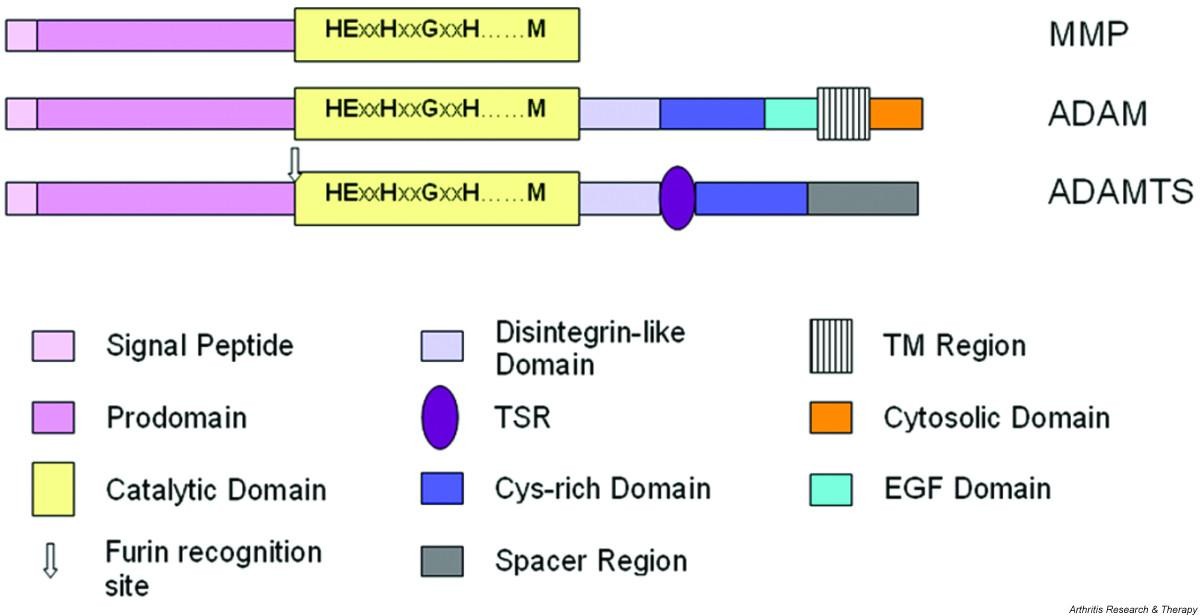
Minimaalisinta kaavamaista domaanien ärjestäytymsitä kuvataan alla. Siinä on MMP, ADAM ja ADAMTS esitettynä domaaneineen.
MMP, metalloproteinaasi
ADAM, eräs disintegriini. ja metalloproteinaasi
ADAMTS, eräs disintegriini ja metalloproteinaasi, jossa on trombospondiinidomeeni (TSR).
Huomioi, että useimmilla MMP-proteinaaseilla on lisäksi C-terminaalisia pidentymiä, joissa on hemopexiinin-kaltaisia domaneja ja fibronektiini II-tyyppisiä domeeneja.
ADAMTS proteinaasit omaavat 0 - 14 kpl trombospondiini tyyppi 1:n kaltaisten motiivien toistoja (TSR) C-terminaali Spacer alueeseen.
Figure 1 Schematic representation of the minimal domain organisation of matrix metalloproteinase (MMP), ADAM (a disintegrin and metalloproteinase) and ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs; for example ADAMTS-4) proteinases. Note that most MMPs possess additional C-terminal extensions containing domains such as hemopexin-like and fibronectin type II domains. ADAMTS possess from 0 to 14 additional thrombospondin type 1-like repeat (TSR)-like motifs C-terminal to the spacer domain.
EGF, epidermal growth factor;
TM, transmembrane.
https://arthritis-research.biomedcentral.com/articles/10.1186/ar1783
ADAMTS- perheen jäsenet ( disintegriini ja metalloproteinaasi, jolla on trombospondiinimotiiveita) unnetaan siitä, että se vaikuttaa kehitykseen, angiogeneesiin, koagulaatioon ja artriitin progredioitumsieen.
Proteinaasina niillä on seuraavia substraatteja:
- von Willebrand-faktorin prekursori, edeltäjäproteiini,
- ECM- komponentit( extrasellularisen matriksin komponentit) kuten prokollageeni, hyalektaanit ( hyaluronaania sitovat proteoglykaanit, aggrekaani niiden joukossa)
- dekoriini
- fibromoduliini
- ruston oligomeerinen matriksiproteiini
ADAMTS-proteinaasien pitoisuudet ja aktiviteetit omaavat monitasoisen säätelyjärjestelmän ja ne säätyvät geeni-ilmentymän kontrollista, mRNA-pleissauksesta, proteiinin prosessoitumisesta ja TIMP- metalloproteinaasien kudosestäjien vaikutuksesta . Ihmisen rustoseulontatutkimukset ovat osoittaneet, että moninaiset ADAMTS-perheen jäsenet saattavat olla tärkeitä sidekudoksen homeostaasissa ja patologiassa.
(Suomennosta tiivistelmästä)
Abstract
- Members of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family are known to influence development, angiogenesis, coagulation and progression of arthritis. As proteinases their substrates include the von Willebrand factor precursor and extracellular matrix components such as procollagen, hyalectans (hyaluronan-binding proteoglycans including aggrecan), decorin, fibromodulin and cartilage oligomeric matrix protein. ADAMTS levels and activities are regulated at multiple levels through the control of gene expression, mRNA splicing, protein processing and inhibition by TIMP (tissue inhibitor of metalloproteinases). A recent screen of human cartilage has shown that multiple members of the ADAMTS family may be important in connective tissue homeostasis and pathology.
ADAMTS-proteinaasien evoluutiosta ja rakenteesta.
ADAMTS evolution and structure
- ADAMTS proteinases were first described in mice by Kuno and colleagues in 1997 [4] and have subsequently been identified in mammals and Caenorhabditis elegans. They form part of subfamily B (adamalysin subfamily), family M12, in clan MA of the metallopeptidases, as defined in the MEROPS database [5, 6] and are structurally and evolutionarily related to the ADAM (a disintegrin and metalloproteinase; also part of the adamalysin subfamily) enzymes and, more distantly, the matrix metalloproteinase (MMP; family M10 in clan MA) enzymes. A comparison of the minimal characteristic domain organisation of these groups of proteinases is shown in Fig. 1.
ADAMTS-geenien koodaamat proteiinit
Ihmiseltä on tunnsitettu 19 eri ADAMTS- geenituotetta. Fylogeneettisiä ja geenianalyysiä on tehty näistä sekvenssien perusteella ja ADAMTS- proteiinit voidaan jakaa yleisesti ottaen neljään alaryhmään, joissa havaitaan myös rakenteellisia ja aktiviteetille ominaisia piirteitä . Tästä on myös kuva 2. KAtalyyttisistä domeeneistä , sekvensseistä , tehty dendrogrammi osoitaa miltei identtisiä domeeneja, mikä osoitaa että katalyyttiset ja niitä edistävät domeenit ovat evoloituneet yhdessä .1. Ensimmäinen ADAMTS-ryhmä käsittää ADAMTS-1,-4,-5,-8,-9,-15, ja -20. ja se jakautuu kahteen alaryhmäänjoista
toisessa on ADAMT-9 ja ADAMTS-20 ja
muut mainitut ovat toisessa alaryhmässä ( ADAMTS-1,-4,-5,-8,-15).
2. Toinen selvästi erotettavissa oleva ADAMTS ryhmä käsittää ADAMTS-2, -3 ja -14.
3, Kolmannessa ryhmässä on pelkästään ADAMTS-13.
4. Neljännessä, hatarammin määritellyssä ryhmässä ovat muut jäljellä olevat. ne ovat edelleen luokiteltu neljään pariin rakenteellisten piirteiden perusteella:
ADAMTS-19 ja -17,
ADAMTS-18- ja 16,
ADAMTS-12 ja -7,
ja ADAMTS -10 ja -6.
On myös julkaistu yksityiskohainen tutkimus ADAMTS-perheen jäsenten välisistä fylogeneettisistä suhteista.
- Nineteen distinct human ADAMTS gene products have been identified. A nearest-neighbour dendrogram constructed (using ClustalW 1.7 [7]) from sequence alignments of the entire protein indicates that human ADAMTS proteins can be broadly divided into four subdivisions, which also seem to share structural characteristics and activities (see Fig. 2 and below). A dendrogram constructed from the sequence alignment of the catalytic domains was almost identical, which implies that the catalytic and ancillary domains evolved together (data not shown). The first of the divisions, consisting of ADAMTS-1, -4, -5, -8, -9, -15 and -20, subdivides into two further groups, one composed of ADAMTS-9 and -20 and the other of ADAMTS-1, -4, -5, -8 and -15. A second, well-defined, subgroup contains ADAMTS-2, -3 and -14. ADAMTS-13 stands alone, and the remaining ADAMTS members form a loosely defined subgroup within which members are further divided into four pairs (ADAMTS-19 and -17, ADAMTS-18 and -16, ADAMTS-12 and -7, and ADAMTS-10 and -6) sharing structural features. A detailed study of the phylogenetic relationship of the ADAMTS family members has recently been published [8].
ADAMTS domain structure
The
signal sequence of ADAMTS proteins is followed by a pro-region of
varying length, but which is unusually short in ADAMTS-13. The
pro-domain of all ADAMTS proteinases contains at least one furin
cleavage consensus motif; it is therefore generally believed that the
zymogen forms of ADAMTS proteinases are cleaved intracellularly and that
secreted proteins are in the mature form. This mechanism of maturation
is supported by studies of ADAMTS-4, which identify an N terminus of F213ASLS in supernatants conditioned by cells transfected with ADAMTS-4, suggesting that the prodomain is efficiently removed in vivo [9].
The same study also demonstrated that purified proADAMTS-4 could be
cleaved by recombinant furin in cell-free experiments. Furin has
recently been shown to interact with the pro-form of ADAMTS-4 and to
co-localise within the trans-Golgi network [10].
Using furin inhibitors and RNA interference techniques, the removal of
the pro-domain was inhibited without affecting secretion, demonstrating
an important role for furin in intracellular processing [10]. The same study also revealed the presence of furin-independent pro-domain processing pathways in some cells.
The catalytic domains of ADAMTS proteinases share a high degree of
similarity and contain the zinc-binding sequence HEXXHXXGXXH, in which
the catalytic zinc is coordinated by the three histidine residues. This
arrangement is facilitated by the conserved glycine, which permits a
tight hairpin loop and enables the third histidine to occupy its correct
position [11, 12].
As in all MMPs and adamalysins, the zinc-binding sequence is followed
at a short distance C-terminally by a conserved methionine residue, an
active-site arrangement that has been termed 'metzincin-type'. This
methionine constitutes the 'Metturn', a tight turn arranged as a
right-handed screw that seems to serve an important function in the
structure of the active site [11].
The catalytic domain is followed by a region with 25 to 45% identity to
the snake venom disintegrins, although it does not contain the cysteine
arrangement of the latter [13].
This domain has therefore been termed disintegrin-like, though there is
currently no published evidence that this ADAMTS domain interacts with
integrins.
Unlike
ADAM proteins, ADAMTS proteinases possess a well-conserved
thrombospondin type 1-like repeat (TSR), homologous to the type I
repeats of thrombospondins 1 and 2 [14], between the disintegrin-like and cysteine-rich domain (CRD). By analogy to thrombospondins 1 and 2 [15],
the central TSR of ADAMTS proteinases is believed to function as a
sulphated glycosaminoglycan-binding domain. The independently expressed
central TSR of murine ADAMTS-1 required 0.46 to 0.66 M NaCl for elution
from a heparin affinity column, indicating that this motif forms a
functional heparin-binding unit [16].
Unlike
ADAM proteins, ADAMTS proteinases possess a well-conserved
thrombospondin type 1-like repeat (TSR), homologous to the type I
repeats of thrombospondins 1 and 2 [14], between the disintegrin-like and cysteine-rich domain (CRD). By analogy to thrombospondins 1 and 2 [15],
the central TSR of ADAMTS proteinases is believed to function as a
sulphated glycosaminoglycan-binding domain. The independently expressed
central TSR of murine ADAMTS-1 required 0.46 to 0.66 M NaCl for elution
from a heparin affinity column, indicating that this motif forms a
functional heparin-binding unit [16].
With
the exception of ADAMTS-4, which terminates after the spacer region,
all ADAMTS proteinases possess between 1 and 14 TSRs C-terminal to the
spacer region (Fig. 2).
The sequence of these additional TSRs is more variable between the
ADAMTS proteinases than is the central TSR, but the independent
expression of the C-terminal TSRs of murine ADAMTS-1 has indicated that
these motifs can form functional heparin-binding units [16].
The TSRs of the C-terminal region are arranged in one, two or three
tandem arrays. Between arrays is either a short linker sequence
(ADAMTS-9 and ADAMTS-20) or a mucin-like domain (ADAMTS-7 and ADAMTS-12)
[17].
Four
additional types of module have been described in the ADAMTS group and
all are present C-terminal to the TSR arrays. ADAMTS-9 and -20 contain a
unique module, also found in the C. elegans ADAMTS GON-1, containing 10 conserved cysteine residues [18].
Several ADAMTS proteinases (-6, -7, -10, -12, -16, -17, -18 and -19)
possess a PLAC (protease and lacunin) domain containing six conserved
cysteine residues, which is found in some pro-protein convertases [19].
A C-terminal extension containing a unique embedded PLAC domain is
present in ADAMTS-2, -3 and -14. Finally, CUB (complement C1r/C1s, Uegf
(EGF-related sea urchin protein) and BMP-1 (bone morphogenic protein-1))
domains are present at the C terminus of ADAMTS-13 [20].
This domain is also present in spermadhesins, tumour necrosis
factor-stimulated gene-6 and the complement proteins C1r, C1s and
mannan-binding lectin-associated serine proteinases, among others [21],
and there is evidence to suggest that these domains mediate
protein-protein interactions with other CUB domain-containing proteins [22, 23].
Inga kommentarer:
Skicka en kommentar